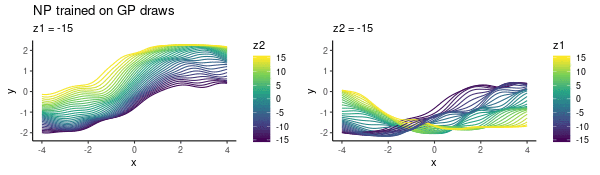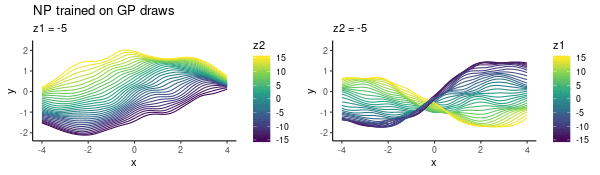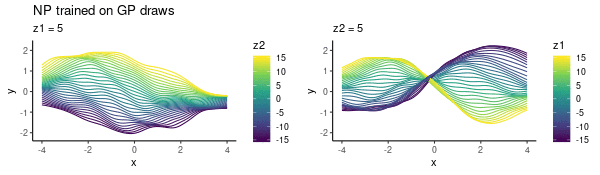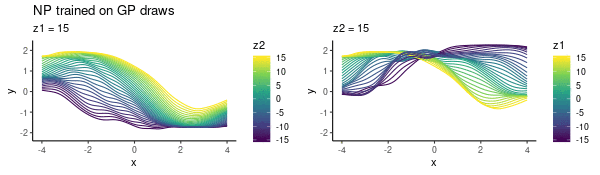To enhance LLM biological reasoning about cell perturbation outcomes, we fine-tune models on synthetic reasoning traces from frontier LLMs, achieving substantial performance gains on PerturbQA.
L Phillips, M Boubnovski Martell, A Misra, J Stoisser, C Prada, R Donovan-Maiye, K Märtens
Arxiv
LangPert combines Large Language Models with numerical methods (kNN) to predict cellular responses to genetic perturbations, overcoming LLMs’ limitations with high-dimensional genomic data while achieving state-of-the-art performance.
K Märtens, M Boubnovski Martell, C Prada, R Donovan-Maiye
MLGenX at ICLR 2025 (selected as Oral Presentation)
Genetic perturbations are key to understanding how genes regulate cell behavior, yet the ability to predict responses to these perturbations remains a significant challenge. While numerous generative models have been developed, most lack the capability to generalize to perturbations not encountered during training. To alleviate this limitation, we introduce a novel methodology that incorporates prior knowledge through embeddings derived from LLMs, effectively informing our predictive models with a deeper biological context.
K Märtens, R Donovan-Maiye, J Ferkinghoff-Borg
Accepted to MLGenX workshop at ICLR 2024
Here, we investigate the capabilities of Multimodal Variational Autoencoders, such as MVAE and MMVAE, to reliably infer private and shared latent factors from multi-omics data. In particular, we highlight a challenging problem setting where modality-specific variation dominates the shared signal. Taking a cross-modal prediction perspective, we discuss and demonstrate limitations of existing models, and propose a modification how to make them more robust to modality-specific variation.
K Märtens, C Yau
Accepted to MLCB (2023)
Due to the black-box nature of VAEs, their utility for healthcare and genomics applications has been limited. Here, we focus on characterising the sources of variation in Conditional VAEs. Our goal is to provide a feature-level variance decomposition, to separate out the marginal additive effects of latent variables z and fixed inputs c from their non-linear interactions. We propose to achieve this through what we call Neural Decomposition - an adaptation of the functional ANOVA decomposition from classical statistics to deep learning models.
K Märtens, C Yau
Accepted to AISTATS (2020)
It would be desirable to construct a joint modelling framework for simultaneous dimensionality reduction and clustering of features. Here, we focus on embedding such capabilities within the Variational Autoencoder (VAE) framework. Specifically, we introduce a probabilistic clustering prior within the VAE decoding model which lets us learn a one-hot basis function representation. For scenarios where not all features are aligned, we develop an extension to handle translation-invariant basis functions.
K Märtens, C Yau
Accepted to AISTATS (2020)
The interpretation of high-dimensional data typically requires the use of dimensionality reduction techniques. However, in many real-world problems the extracted low-dimensional representations may not be sufficient to aid interpretation on their own, and it would be desirable to interpret the model in terms of the original features themselves. Our goal is to characterise how feature-level variation depends on latent low-dimensional representations, external covariates, and non-linear interactions between the two. Here, we propose to achieve this through a structured kernel decomposition in a hybrid Gaussian Process model which we call the Covariate Gaussian Process Latent Variable Model (c-GPLVM).
K Märtens, K Campbell, C Yau
Accepted to ICML (2019)
Here, we introduce an augmented ensemble MCMC technique to improve on existing poorly mixing samplers for factorial HMMs. This is achieved by combining parallel tempering and an auxiliary variable scheme to exchange information between the chains in an efficient way.
K Märtens, MK Titsias, C Yau
Accepted to AISTATS (2019)
Here, we explore whether accurate complex trait predictions can be achieved in practice. Using a genome sequenced population of ∼7,000 yeast strains of high but varying relatedness, we consider a variety of models for predicting growth traits from various sources of information (family information, genetic variants, and growth in other environments).
K Märtens, J Hallin, J Warringer, G Liti, L Parts
In Nature communications (2016)
Here, we propose a statistical method for identifying differentially methylated regions, based on the minimum description length (MDL) principle. Our method is available as an R package.
R Kolde*, K Märtens*, K Lokk, S Laur, J Vilo
In Bioinformatics (2016)